RESEARCH ROUNDUP
THE FUTURE OF HUMAN HEALING LIES IN THE BRAIN OF A STARFISH
The incredible benefits of stem cell therapy have been widely known for decades. It can alleviate the pain of arthritis and help patients heal exponentially faster after surgery. But stem cell therapies are prohibitively expensive — to the tune of $100,000 to $200,000 per patient. Because of the exorbitant cost, these life-changing treatments are far out of range of the vast majority of those who need them. But thanks to Professor and Head of Biological Sciences Veronica Hinman and Professor of Chemical Enginering Kris Noel Dahl, stem cell therapy is about to get a whole lot cheaper. And the key to it all lies in the incredible regenerative powers of starfish.
“Many species of animals, including starfish, have extraordinary capacities to regenerate and can reform all lost body parts following traumatic injury,” says Hinman. “This capacity derives in part from their natural abilities to dedifferentiate and reprogram cells. Understanding how cellular reprogramming occurs in nature has great potential for scientists to translate these findings to human cell cultures to identify cheap and robust ways to generate stem cells.”
Stem cells are the body’s raw material, able to become any type of cell the body needs: muscle cells, skin cells, blood cells and more. In humans, when stem cells differentiate into these other cells, they are unable to change back, or dedifferentiate. And they most definitely can’t turn into a different kind of cell. But this is not the case with starfish. Notably, if a starfish loses one of its arms, it can grow it back just the same. This is due to the starfish’s unique cells, which can dedifferentiate themselves from skin or muscle cells back into stem cells. While this regenerative capability in itself is incredible, Hinman’s preliminary research has shown an even more incredible ability.
’“In the larval state, starfish have a distinctive head that contains their brain,” says Dahl. “If the head is removed or damaged, the differentiated cells that are definitely not neural cells will dedifferentiate, crawl up to the head region, and regrow into neurons. To not only do this in the larval state, but to regrow something as complex as a brain — this is an amazing regenerative capability.”
Supported by funding from the DSF Charitable Foundation block grant program, Hinman and Dahl are working to understand just what in the starfish causes their cells to do this. While Hinman is focused on the fundamental science, Dahl’s lab is delving into the structure of the cells, cell crawling and the biomechanics of cellular regeneration.
The Hinman lab uses the larvae of starfish to understand the genomic control of development and cell fate decisions. They have developed tools to engineer transgenic larvae, measure gene expression and chromatin changes, and track cells during specification. With these tools, Hinman and her team have shown that following amputation, cells at the wound site turn on pluripotency factors and undergo cell fate reprogramming to regenerate lost tissue.
“While regenerative medicine is great, there’s still a lack of understanding of the fundamentals that govern how cells respecify themselves,” says Dahl. “The hope is that by studying a model organism like the starfish and combining what we learn with our knowledge of human stem cells, we can use comparative genomics to understand the gene expression that allows starfish cells to respecify their programming.”
To do this, Dahl is creating an artificial model of the starfish’s larval system to map the cells as they crawl to their new destination. With this artificial model, Dahl and her team can manipulate the chemical and mechanical factors that exist in the starfish embryo, blocking them one at a time until they find exactly what it is that tells the cells to dedifferentiate back into stem cells, crawl up to the brain region and become neuronal tissue. Once this factor has been isolated, the goal is to then be able to apply it to human cells, to tell those cells to dedifferentiate so they can become whatever the patient needs.
Current therapies require stem cells to be harvested from a patient, then cultured over the course of days, in order to have enough to be reinjected back into the patient to help speed healing. But with this new method, cells could be taken from any part of the body, dedifferentiated back into stem cells, then re-differentiated into therapeutic cells. This could make the process of preparing stem cell therapies faster, easier and, most importantly, cheaper.
“If you could reduce stem cell therapy from $200,000 to $1,000 — it would touch nearly every person’s life,” says Dahl. “Surgeons could include a stem cell injection with every major or minor surgery, helping patients heal 100 times faster. It’s quick healing, it’s reduced scarring: this could be like penicillin. I see it becoming the standard of care in the next 10 years.”
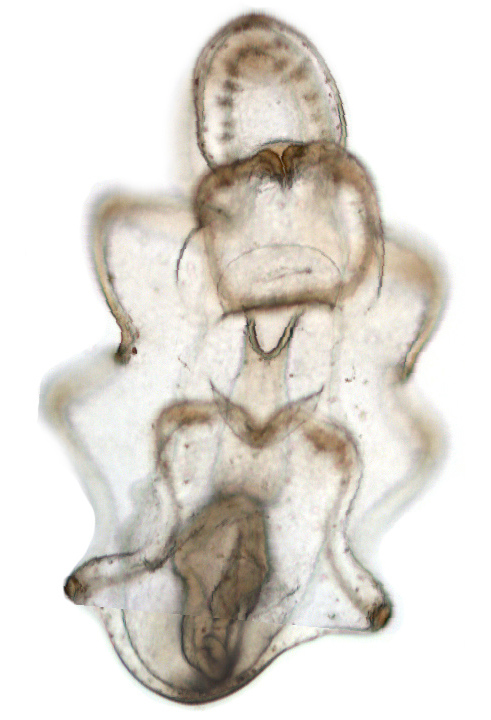
Starfish embryo (Patiria miniata) twelve days after fertilization. (Hinman Lab)

Charles Ettensohn
CHARLES ETTENSOHN RECEIVES GRANT TO STUDY THE GENOME
Professor of Biological Sciences Charles Ettensohn has received a $1.2 million grant from the National Science Foundation to support his research into how an organism’s genome regulates the development of its body from scratch.
“Remarkably, the entire blueprint for embryonic development is contained in the DNA sequence of the genome,” Ettensohn said. “A central challenge of modern biology is to explain how this genomic information is ‘decoded’ to control the process of embryogenesis.”
In particular, Ettensohn is interested in understanding an organism’s gene regulatory network, a collection of molecules that control how and when genes are expressed. This network takes on vital importance during the development of an embryo when the structure of an organism’s developing body is mapped out. Using sea urchins as a model organism, Ettensohn plans to study how this network works over the course of an embryo’s development, and how it can go wrong.
“This grant allows us to use genomics-based tools to dissect the structure and function of a model developmental gene regulatory network as a paradigm for understanding how the genome of an organism controls its anatomy,” Ettensohn said.
USING CHEMISTRY AS A LENS TO AID IN DIAGNOSING DISEASES
A team of researchers led by Assistant Professor of Biological Sciences Yongxin (Leon) Zhao has improved a powerful cellular magnification technique, making it more practical for use in clinical settings.
“For almost 200 years, pathologists have used optical microscopes to examine biopsies from patients,” said Zhao. However, the diffraction limit of light means that minute, but important, details in tissue samples can’t be seen with even the best conventional optical microscopes.
More powerful techniques and devices, such as super-resolution optical microscopes and electron microscopes, do exist; however, they are expensive and technically complex and therefore not available to most pathologists. To get around this issue, Zhao and collaborators have pioneered a technique known as Expansion Pathology, which can magnify the details of human tissues by physically making them larger. This allows normal microscopic equipment to see far more detail than ordinarily possible.
In a new collaborative paper published in the journal Nature Protocols, Zhao and his colleagues at the Massachusetts Institute of Technology and Harvard Medical School laid out step-by-step how this new field of Expansion Pathology can enable pathologists to inexpensively and rapidly track even the subtlest changes in human tissue.
Zhao likens Expansion Pathology to looking at the Earth with an ordinary backyard telescope. You may be able to see some features of the planet but nothing in close detail. Now imagine if you were able to literally stretch the Earth and everything on it in all directions equally, expanding it in size. Suddenly, with that same ordinary telescope you would be able to see houses, trees and even people.
The technique works by chemically embedding the specimen in a hydrogel, which is a substance made of cross-linking polymer chains that expand when they come into contact with water. Perhaps best known for being used in baby diapers, these hydrogels tug on the tissue from all directions and stretch it to a larger size.
“We use chemistry as a lens to further magnify the details of the pathological samples,” Zhao explained.
Previous iterations of this technique would take around 30 hours to properly expand and image samples, but in their newest paper Zhao and his collaborators introduce an improved version called Rapid Expansion Pathology that takes only four to eight hours to use, which significantly lowers the barrier for adaptation in the fast-paced clinical setting.
“This technique can be applicable to a wide range of tissues or questions that you want to work with and it can be further empowered by machine learning,” Zhao said. “Basically, now the limitation is what kind of questions do you want to ask that can benefit from this kind of technology.”
On that end, Zhao is now working with pathologists to conduct new clinical studies enabled by Expansion Pathology that may one day foster new diagnostics.
“It’s not just a technique — it’s a new possibility to open up the field,” Zhao said.

Specimen expanding after being chemically embedded in a hydrogel. (Zhao Lab)

Aryn Gittis
NEW TECHNIQUE HELPS RESEARCHERS PREDICT MOTOR DYSFUNCTION
Research from the lab of Aryn Gittis, associate professor of biological sciences and Neuroscience Institute member, found a new way to predict problems related to Parkinson’s disease. In a new paper published in the Journal of Neurophysiology, Gittis and her team showed that delta oscillations, a type of brain wave, are excellent indicators of dopamine loss, which leads to motor dysfunction.
The researchers developed a new technique to detect spike oscillations and found that they are better at predicting the severity of motor dysfunction than any other physiological metric. This work represents how collaborations across the boundaries of mathematics and neuroscience can advance our understanding of how patterns of neural activity change in disease.
Gittis and her team hope to eventually develop interventions that could help Parkinson’s disease patients.
“We hope this work will spark a renewed interest in delta oscillations in Parkinson’s disease that could be monitored in patients for therapeutic or diagnostic purposes,” she said.
The lead author on the paper was Timothy Whalen, a joint Ph.D. student in the Gittis lab (CMU) and Rubin lab at the University of Pittsburgh. Other co-authors on the study were Amanda M. Willard, a former Gittis lab member, and Jonathan E. Rubin from the Pitt department of mathematics.
HUAIYING ZHANG RECEIVES NIH GRANT TO STUDY MECHANISMS OF CANCER GROWTH
Assistant Professor of Biological Sciences and Chemical Engineering Huaiying Zhang has received a five-year, $2.5 million grant from the National Institutes of Health to study how abnormalities in the nuclear organization of a type of cancer cell affects their development.
Typically, a human cell can only divide a certain number of times before it dies, with this process being regulated by the length of its telomeres, a sequence of nucleotides at the end of the cell’s chromosomes. Most cancers can get around this restriction by reactivating the enzyme telomerase to maintain the telomeres’ length and allow them to keep dividing. But 10% to 15% of cancers use a different method to keep dividing infinitely called alternative lengthening of telomeres (ALT).
ALT cancer cells typically show many abnormalities in the layout of their nuclei, and these distinctions are often used in diagnosing this type of cancer. Scientists have theorized that targeting these abnormalities could be a useful form of cancer treatment since, unlike more general treatments such as chemotherapy, these targeted therapeutics would avoid harming normal cells that lack the abnormalities.
Scientists still have little understanding of how exactly these abnormal cell features develop and contribute to cancer growth. Zhang has proposed a liquid-liquid phase separation model for the abnormalities’ assembly and function. To investigate that hypothesis, Zhang will collaborate with Associate Professor Ulrike Endesfelder from the Department of Physics, Professor Kris Dahl from the College of Engineering and Associate Professor Roderick O’Sullivan from the University of Pittsburgh’s Hillman Cancer Center.
“It’s so exciting as a new professor to be able to bring together a group of people to do meaningful work on cancer research,” Zhang said. “Our results can inspire novel strategies for the detection and treatment of ALT cancer targeting its unique nuclear organization.”

Huaiying Zhang
