Research Roundup
New Fluorescence Imaging Method Reveals Neural Circuits Altered By Learning
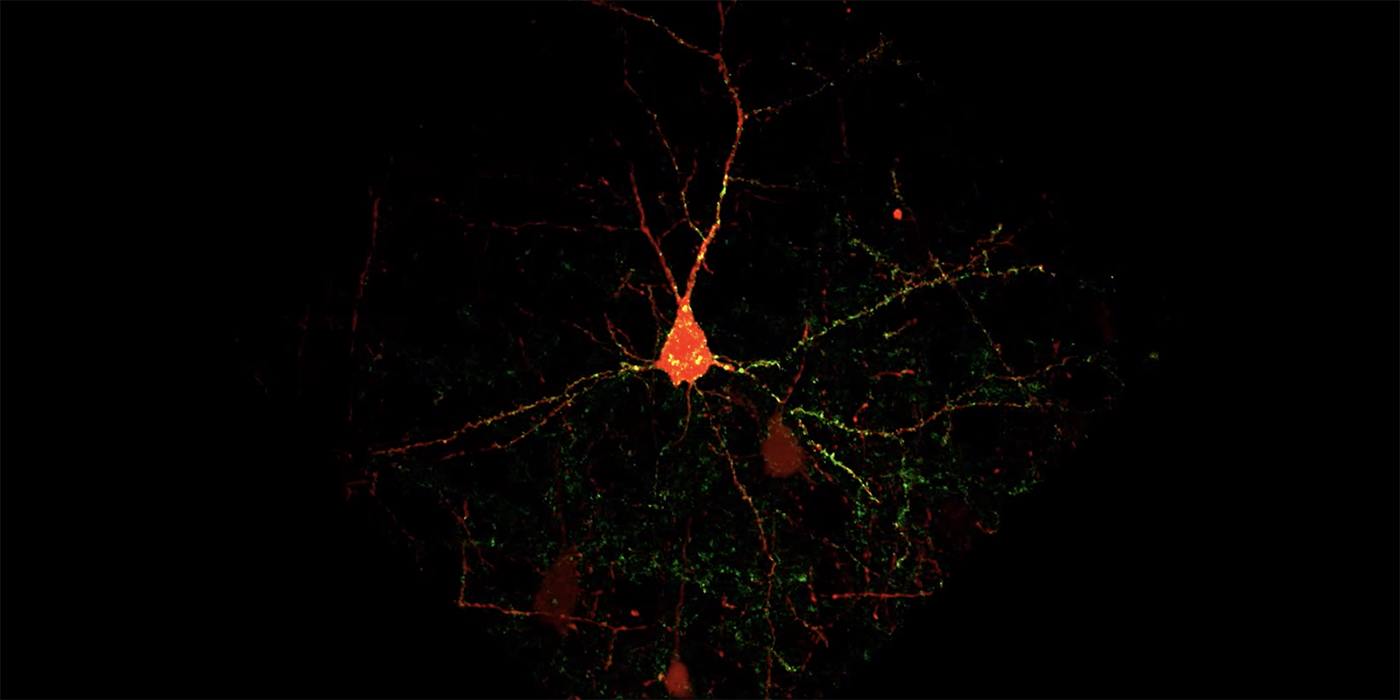
Carnegie Mellon University researchers have developed a new fluorescence-based method for detecting synaptic connections between specific types of neurons, allowing them to detect and quantify subtle structural changes that take place during learning. Using molecular genetic tools for cell-type specific labeling of synapses, the researchers could identify which connections were changing as mice learned a sensory association task. The research was published in the Journal of Neuroscience.
“There are trillions of synapses in the brain, and finding ones that have been modified during learning is a classic needle-in-the-haystack problem,” said Alison Barth, the Maxwell H. and Gloria C. Connan Professor of Biological Sciences and member of Carnegie Mellon’s Neuroscience Institute. “This new method allows us to detect very subtle shifts in the distribution of synapse size to see how specific synapses are modified as animals are mastering a task.”
The brain changes with learning and experience, an ability known as plasticity. Much of that change happens at synapses, the small gaps between neurons where they pass messages to communicate. Although it has long been appreciated that synaptic plasticity is critical for learning, figuring out which synapses are modified has been a daunting task.
Neuroscientists typically study synaptic plasticity by measuring neurons’ electrical response to a stimulus like a touch or sound, which can indicate that a synaptic connection has been strengthened or weakened. While existing electrophysiological recording techniques are good at indicating when a neuron fires and how big the response is, they are not good at showing where the response occurred. The where is critical.
“For a very long time, people have been studying how synapses are changing, but they had no idea how to relate synapse dynamics to information flow without knowing the presynaptic partner,” Barth said. “A synapse is a conversation between partners — listening to half of the conversation can’t tell you what is being discussed. It really matters where these synapses come from and where they are going to.”
In the current study, Barth and her colleagues examined how a specific neural circuit — a series of connected neurons that send signals from one part of the brain to another — changes as mice learn to associate a whisker stimulus with a water reward. The researchers modified existing multicolor, fluorescence-based tags to label the components of the circuit: axons from neurons originating in the thalamus (a part of the brain sensitive to context and rewards), dendrites of neurons in the somatosensory cortex (which processes sensations from the body), and the synapses where the two types of neurons meet and communicate.
The research team, led by postdoctoral associate Ajit Ray, has spent significant time recording from these neurons, so they knew some of the synapses were getting larger in response to learning. With their fluorescent imaging method, the team was able to expand their scope to visualize tens of thousands of synapses, which is orders of magnitude more synapses than can be measured with current recording techniques. The new approach enabled them to see small differences that otherwise may go unnoticed.
The researchers discovered that these synapses are modified in response to learning, but not where they expected. Barth said they had good reason to think that the neurons in layer 1 of the somatosensory cortex were changing, but it turned out that they weren’t.
“They were actually changing deeper in the brain,” Barth said. “But nobody had ever looked before because there wasn’t any way to see them.”
The researchers also discovered that the synaptic changes are temporary. Once the animal becomes an expert at the task, the modified synapses revert to their original state, suggesting that there is a transient enhancement of information processing in the cerebral cortex while learning is happening.
“There’s a whole cottage industry of people who are trying to figure out where in the brain memories lie. If you have a visual or a touch memory, the obvious place to look is in the part of the brain that encodes vision or touch. And yet, if those changes are just very brief, there’s a glimmer of change and then they disappear, then they may be stored somewhere else,” Barth said. “I think what we are finding is that the somatosensory cortex helps you learn it, but it doesn’t necessarily help you remember it later.”
Barth, Ajit and the research team are using their imaging technique now to look at other neural pathways.
“We think this technique can be broadly applied to many types of synapses and to other pathways that might be altered during learning as well as pathways that might be altered in different disease states,” said Barth.
She acknowledges that while their fluorescence approach doesn’t offer the type of high-resolution imaging that electron microscopy provides, it does quickly generate huge amounts of fine-scale anatomical data that offers insight into how the brain works.
“Our tool isn’t perfect, but it’s still good enough to tell us things we didn’t know before. And it is a very democratic method,” Barth said. “There are thousands of labs across the country that have what they need to be able to do it. The bottom line is that we can use it to make new discoveries, so that’s a huge advance.”
Additional study authors include: Joseph Christian, Matthew Mosso, and Eunsol Park at Carnegie Mellon, and Waja Wegner and Katrin Willig at the Center for Nanoscale Microscopy and Molecular Physiology of the Brain at the University Medical Center Göttingen and the Max Planck Institute for Multidisciplinary Sciences in Germany. The study was supported by the Carnegie Mellon Neuroscience Institute-Indian Institute of Science Fellowship Program, the National Institutes of Health and the Max Planck Institute for Multidisciplinary Sciences.
■ Amy Pavlak Laird
New Expansion Microscopy Methods Magnify Research’s Impact
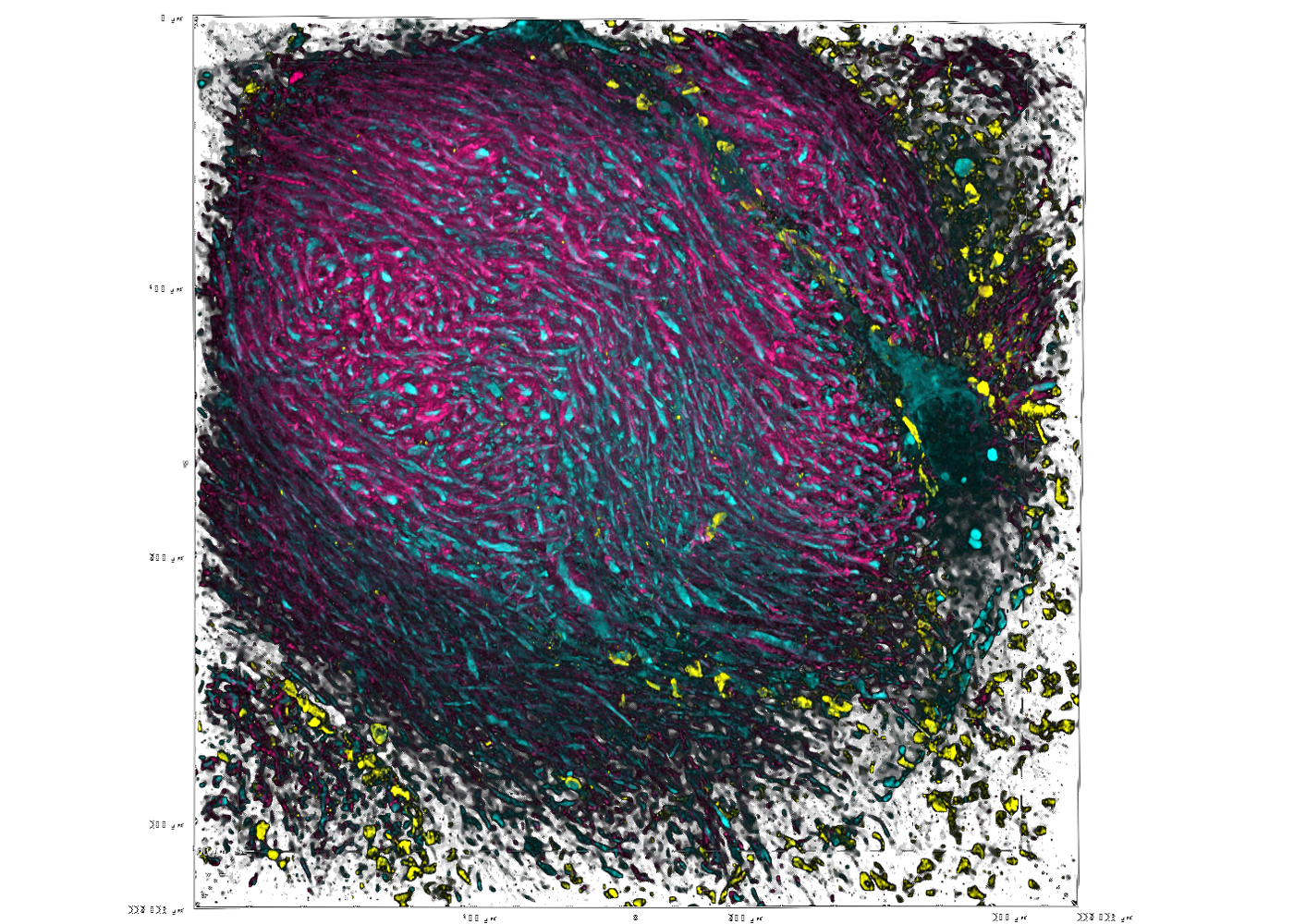
Unprecedented views of the interior of cells and other nanoscale structures are now possible thanks to innovations in expansion microscopy. The advancements could help provide future insight into neuroscience, pathology, and many other biological and medical fields.
In the paper “Magnify is a universal molecular anchoring strategy for expansion microscopy” published Jan. 2 in the journal Nature Biotechnology, collaborators from Carnegie Mellon University, the University of Pittsburgh and Brown University describe new protocols for dubbed Magnify.
“Magnify can be a potent and accessible tool for the biotechnology community,” said Yongxin (Leon) Zhao, the Eberly Family Career Development Associate Professor of Biological Sciences.
Zhao’s Biophotonics Lab is a leader in the field of enabling super-resolution imaging of biological samples through physically expanding samples in a process known as expansion microscopy. Through the process, samples are embedded in a swellable hydrogel that homogenously expands to increase the distance between molecules allowing them to be observed in greater resolution. This allows nanoscale biological structures that previously only could be viewed using expensive high-resolution imaging techniques to be seen with standard microscopy tools.
Magnify is a variant of expansion microscopy that allows researchers to use a new hydrogel formula, invented by Zhao’s team, that retains a spectrum of biomolecules, offers a broader application to a variety of tissues, and increases the expansion rate up to 11 times linearly or ~1,300 folds of the original volume.
“We overcame some of the longstanding challenges of expansion microscopy,” Zhao said. “One of the main selling points for Magnify is the universal strategy to keep the tissue’s biomolecules, including proteins, nucleic acids and carbohydrates, within the expanded sample.”
Zhao said that keeping different biological components intact matters because previous protocols required eliminating many various biomolecules that held tissues together. But these molecules could contain valuable information for researchers.
“In the past, to make cells really expandable, you need to use enzymes to digest proteins, so in the end, you had an empty gel with labels that indicate the location of the protein of interest,” he said. With the new method, the molecules are kept intact, and multiple types of biomolecules can be labeled in a single sample.
“Before, it was like having single-choice questions. If you want to label proteins, that would be the version one protocol. If you want to label nuclei, then that would be a different version,” Zhao said. “With Magnify, you can pick multiple items to label, such as proteins, lipids and carbohydrates, and image them together.”
Lab researchers Aleksandra Klimas, a postdoctoral researcher, and Brendan Gallagher, a doctoral student, were first co-authors on the paper.
“This is an accessible way to image specimens in high resolution,” Klimas said. “Traditionally, you need expensive equipment and specific reagents and training. However, this method is broadly applicable to many types of sample preparations and can be viewed with standard microscopes that you would have in a biology laboratory.”
Gallagher, who has a background in neuroscience, said their goal was to make the protocols as compatible as possible for researchers.
“One of the key concepts that we tried to keep in mind was to meet researchers where they are and have them change as few things in their protocols as possible,” Gallagher said. “It works with different tissue types, fixation methods and even tissue that has been preserved and stored. It is very flexible, in that you don’t necessarily need to redesign experiments with Magnify in mind completely; it will work with what you have already.”
Xi (Charlie) Ren, assistant professor of biomedical engineering at Carnegie Mellon, studies the lung tissue and how to model its morphogenesis and pathogenesis. Part of his research involves researching the motile cilia that function to clear mucus in the human conducting airway. At 200 nanometers in diameter and just a few micrometers in length, the structures are too small to see without time-intensive technology such as electron microscopy. Working in collaboration with Zhao’s lab, Ren’s team developed and delivered lung organoid models with specific defects in cilia ultrastructure and function to validate the ability of Magnify to visualize clinically relevant cilia pathology.
“With the latest Magnify techniques, we can expand those lung tissues and start to see some ultrastructure of the motile cilia even with a regular microscope, and this will expedite both basic and clinical investigations” he said.
The researchers also were able to view defects in cilia in patient-specific lung cells known to have genetic mutations.
The hydrogel used in Magnify and developed in the Zhao lab is more robust than its predecessor, which was very fragile, causing breaks during the process.
“We are hoping to develop this technology to make it more accessible to the community,” he said. “There are different directions this can go. There’s a lot of interest in using this kind of tissue expansion technology for basic science.”
Alison Barth, the Maxwell H. and Gloria C. Connan Professor in the Life Sciences at Carnegie Mellon, studies synaptic connectivity during learning. She said the broad applications provided by the new methods will be a boon for researchers.
“The brain is a great place to take advantage of these super-resolution techniques,” said Barth, who collaborates with the Zhao Lab on several studies. “Microscopy methods will be beneficial for synaptic phenotyping and analysis across different brain conditions.
“One of the major advances in this paper is the method’s ability to work on many different types of tissue specimens.”
Additional study authors include Piyumi Wijesekara, Emma F. DiBernardo, Zhangyu Cheng of Carnegie Mellon; Sinda Fekir and Christopher I. Moore of Brown University; Donna B. Stolz of Pitt; Franca Cambi of Pitt and Veterans Administration; and Steven L Brody and Amjad Horani of Washington University.
This work was supported by Carnegie Mellon, the Kaufman Foundation, and the DSF Charitable Foundation, U.S. Department of Defense (VR190139), the National Institutes of Health (DP2 OD025926-01 and NIH RF1 MH114103), Air Force Office of Scientific Research (FA9550-19-1-13022629), NeuroNex (GR5260228.1001) and Brown University.
■ Heidi Opdyke
Researchers Nanoprint Electrodes for Customized Treatments
The collaboration combines the expertise of Rahul Panat, associate professor of mechanical engineering, and Eric Yttri, assistant professor of biological sciences. The team applied the newest microfabrication technique, Aerosol Jet 3D printing, to produce arrays that solved the major design barriers of other brain computer interface (BCI) arrays. The findings were published in Science Advances.
“Aerosol Jet 3D printing offered three major advantages,” Panat said. “Users are able to customize their MEAs to fit particular needs; the MEAs can work in three dimensions in the brain; and the density of the MEA is increased and therefore more robust.”
MEA-based BCIs connect neurons with external electronics to monitor or stimulate brain activity. They are used in applications like neuroprosthetic devices, artificial limbs, and visual implants to transport information from the brain to extremities that have lost functionality. BCIs also have potential applications in treating neurological diseases such as epilepsy, depression, and obsessive-compulsive disorder. However, existing devices have limitations such as they only record on a two-dimensional plane.
The most important aspect of an MEA is its three-dimensional sampling ability, which is limited by the density of microelectrodes in the array and the ability to position these arrays in the precise spot one wants to sense. Modern MEA manufacturing techniques have made tremendous advances regarding the density of these microelectrode arrays. Adding the third dimension significantly increases the sampling ability of the arrays. In addition, custom-made MEAs for each specific application allow for more accurate and higher-fidelity readings.
Higher-quality MEAs are in demand. MEAs used for controlling virtual actions on a computer or complex limb movements are running up on limitations of the current technology. More advanced applications require MEAs that are customized to each individual and are much higher fidelity than what is currently available.
“Within a matter of days, we can now produce a precision medicine device tailored to a patient or experimenter’s needs,” said Yttri, co-senior author of the study.
Panat predicts that it may take five years to see human testing, and even longer to see commercial use. The team is excited to get this successful process out to other researchers in the field to begin testing a wide variety of applications.
A patent on the CMU Array architecture and manufacturing method is pending. The next step, Panat said, is to work with the National Institutes of Health (NIH) and other business partners to get these findings into other labs as quickly as possible and apply for funding that would commercialize this technology.
The research is funded by the NIH’s Brain Research Through Advancing Innovation Neurotechnologies (BRAIN) Initiative.
Neuroscientists Gain New Understanding of Neural Pathway
In a paper published in Neuron, Aryn Gittis and colleagues present new information about a neural pathway in the basal ganglia, a part of the brain important for skill learning, habit formation and motor control. The paper contradicts the model that has guided researchers’ understanding of motor learning for 30 years.
Gittis, professor of biological sciences, said there are two common theories about what happens in the basal ganglia.
“One school of thought is that the basal ganglia is for motor control — how you stop and go, the gas and brake pedals of the brain,” she explained. “Others think it is involved in learning and shaping movement. If a behavior leads to a reward, we do it more. If it prevents a reward, we stop doing that.”
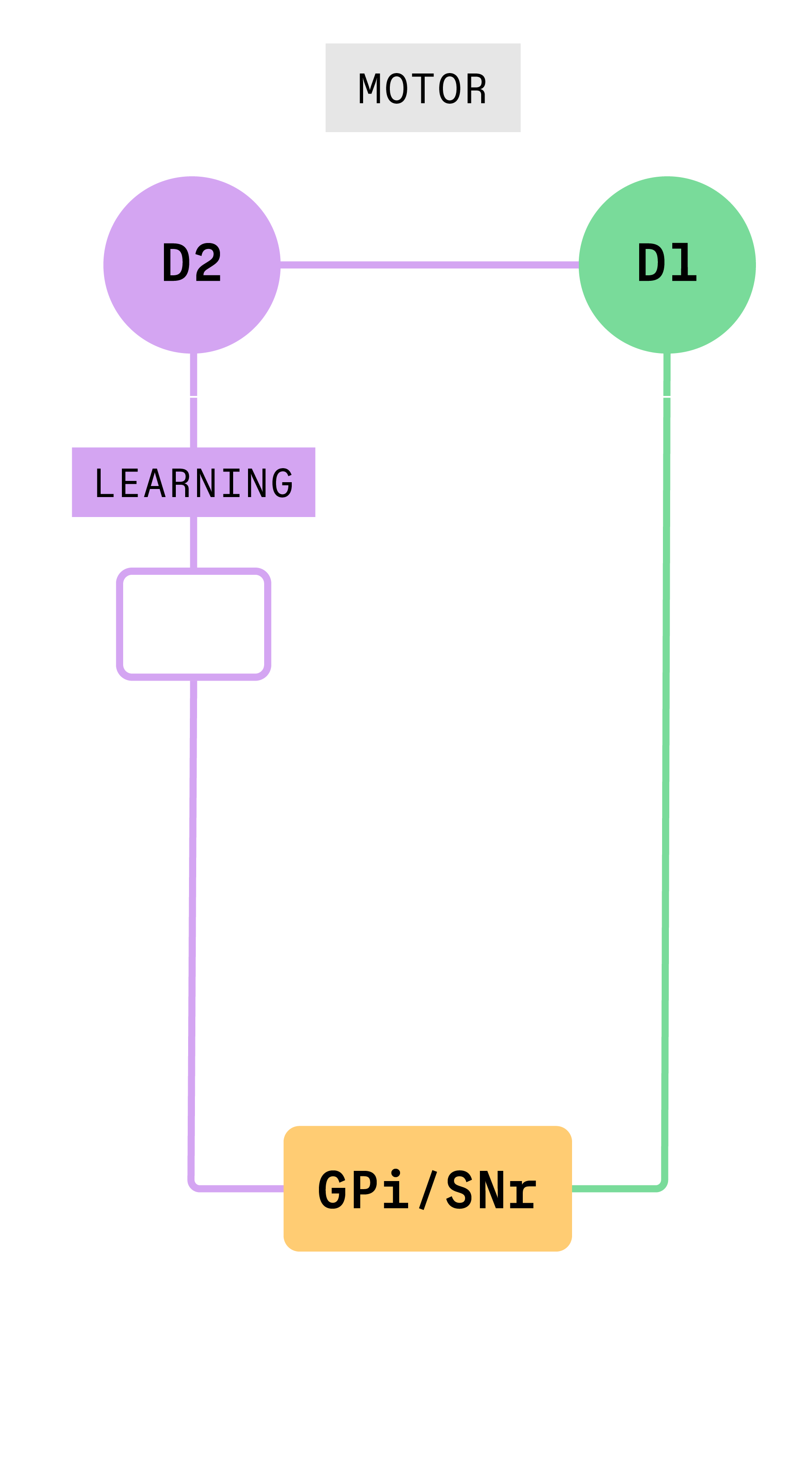
In previous work, researchers showed that activating a specific pathway in the basal ganglia seemed to stop movement in animals, supporting the first theory. To understand why, Gittis and her team took a close look at what happens when specific cells in that pathway, called the “indirect pathway,” are activated.
“We show that when you activate those neurons, you’re actually activating several pathways simultaneously,” Gittis said.
Gittis and her team found that this simultaneous activation of multiple pathways had caused previous researchers to draw the wrong conclusions about the function of the indirect pathway. Stimulating neurons at the beginning of the indirect pathway, D2 neurons, activate two other groups of cells, the globus pallidus (the next cell group in the indirect pathway) and D1 neurons which are part of a different neural pathway. It turned out that the motor effects produced by D2 stimulation that had been attributed to the indirect pathway were actually due to the spurious co-activation of D1 neurons which control motor function.
When experimental conditions were controlled to only activate the indirect pathway, Gittis and colleagues found that this pathway drives learning.
“Rather than driving motor control or directly affecting motor control, it’s shaping behavioral selection,” Gittis said.
Since the pathway affects learning and behavior, it could serve as a target for compulsive behavior treatments in the future. When the indirect pathway is activated, it makes a behavior that was going to happen significantly less likely to happen in the short term. If the same pathway were activated in a person with a compulsive behavior, it could make them less likely to perform the specific compulsive behavior while not blocking all motor function.
Gittis said that this discovery could open doors to additional research, and is proud that her team was able to question long-standing theories.
“You need to question assumptions and really go directly to the data that has led to that assumption. Oftentimes there are gaps that actually hold the key to the discovery,” she said.
■ Caroline Sheedy
McManus Lab Findings Upend Previous Research on Key Leukemia Gene

C. Joel McManus
During the COVID-19 pandemic, the researchers in Associate Professor of Biological Sciences Joel McManus’s lab were finding new ways to further their work when they couldn’t come into the lab due to social distancing restrictions.
With the lab closed, they connected by Zoom, where they discussed published reports from other research groups related to the lab’s work studying how mRNA produces proteins for gene expression. One paper they discussed involved an unusual RNA that was thought to control protein expression through an exotic mechanism. The RNA supposedly folded into a structure that drove the expression of a developmentally critical gene called Hoxa9. But McManus’s team noticed that the data and the conclusions just didn’t seem to match. The structure could be mutated without affecting protein expression.
McManus Lab Research Associate Gemma May then went to a genome browser to compare the Hoxa9 gene annotation with its mRNA sequence. Genome annotations are like a book’s index that can guide researchers to the area of a genome that is responsible for certain functions. She found the annotation didn’t make sense. The part of the gene’s mRNA that was supposed to contain the regulatory structure wasn’t being made. This was particularly important, because Hoxa9 has important roles in development and childhood acute myeloid leukemia.
“The genome annotation says that ‘this gene is located on a specific chromosome and starts in one place and ends in another,’” said McManus. “But Gemma noticed that the annotation was wrong, such that the gene was actually shorter than the public annotation. This meant that the part of the gene that was thought to control protein synthesis really just controlled RNA synthesis.”
McManus said that his group didn’t originally do much with their discovery, until several new reports emerged touting that over a hundred genes contained similar unusual regulatory sequences and, as a result, shared this exotic way of translating proteins. Biological Sciences graduate student Christina Akirtava then set out to check this larger set of genes for evidence of similar annotation errors using computational analyses. At the same time, May tested the regulatory sequences of these genes to see if they did in fact control RNA synthesis in laboratory cells. Together, they found compelling evidence that the published research had been misinterpreted.
“Mouse studies suggested that the gene was being produced by an unusual mechanism where ribosomes bind directly to the inside of the mRNA,” said McManus. “And this was really exciting; it changed the way people thought about how mRNA are translated. But then my lab did some control experiments, and the findings weren’t supported.”
McManus’ group found that Hoxa9 started at a much different position than was recorded in the genome annotation. Part of the gene that was purported to be part of the RNA wasn’t — it was part of the DNA. Their findings, along with those of other labs working on the same problem, overturned at least five high-profile published studies.
“This doesn’t typically happen in our field, where a new model is accepted, then proven to not be what it had claimed to be. We call it paradigm unshifting,” said McManus. The result could have a significant impact on current research on Hoxa9 and efforts by pharmaceutical companies to use the gene to develop new treatments for leukemia. The work also brings into question the accuracy of genome annotations.
“There are big errors in the mouse genome annotation and in the human genome annotation,” said McManus. “The main goal for us was to help other researchers know about these issues so that these problems don’t reoccur in the future.”
■ Jocelyn Duffy