Research Roundup
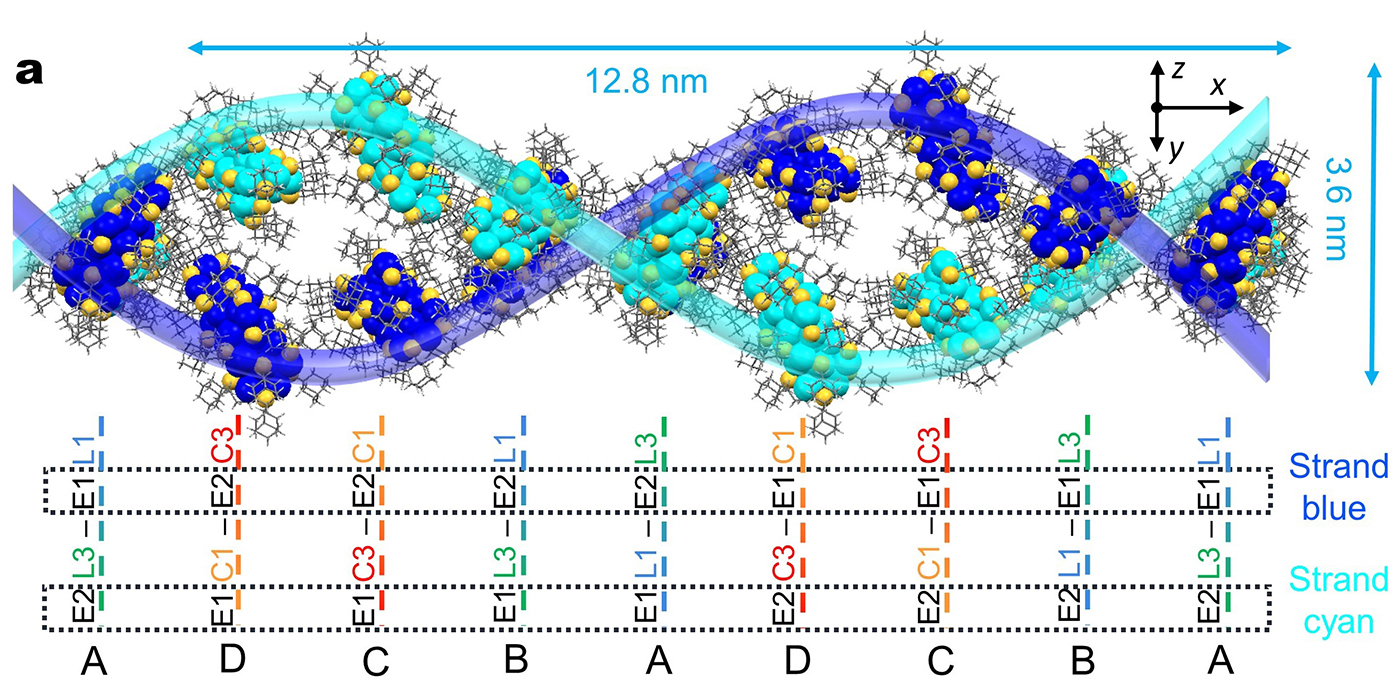
Double-helical assembly of Au29(SAdm)19 nanoclusters in supercrystals
Carnegie Mellon Chemists Create DNA-style Helical Inorganic Supercrystals
Carnegie Mellon University chemists synthesized complex gold supercrystals with arrangements resembling the iconic helical structure of DNA. The work, published in the journal Nature, highlights new promise for the precise design and engineering of supercrystals.
One can think of supercrystals like LEGO blocks, said chemistry Ph.D. candidate Yingwei Li, who was lead author on the study. Li works in the research group of Professor of Chemistry Rongchao Jin, who has pioneered the synthesis of gold nanoparticles with precise numbers of atoms. In this latest study from the group, Li and her collaborators brought Jin’s approach to supercrystals. These macroscopic crystals are built piece-by-piece out of nanoparticles or nanoclusters, allowing for the possibility of creating meticulously tailored materials with a variety of different properties.
The researchers synthesized gold nanoclusters in two varieties based on a dimeric structure, which comprises two roughly spherical parts combined. The homodimeric nanoclusters had two largely identical component parts, and these nanoclusters arranged themselves into familiar layered superstructures, Li said. But the heterodimeric nanoclusters behaved far differently.
“To our surprise, the heterodimeric nanoclusters self-assemble into double- and quadruple-helical superstructures,” Li said.
The helical structure of DNA has long been a model for constructing particle assemblies, the researchers noted, but getting inorganic nanoclusters to form such structures has been challenging. A complex series of atomic interactions and bonding help nucleotides form DNA helices, a feature purely inorganic particles are not able to use.
However, by carefully crystallizing heterodimeric nanoclusters, Li and her collaborators were able to leverage the natural attractions between molecules called Van der Waals interactions to create the inorganic helices.
“Our work demonstrates a new principle of constructing supercrystal materials, and the resulting exquisite supercrystals may lead to other discoveries of functionality,” Li said.
Li and her co-authors are now working on closely studying the properties of these newly created supercrystals and how they could potentially be used.
Other authors on this study included Tatsuya Higaki of Carnegie Mellon, Meng Zhou and He Wang of the University of Miami and Yongbo Song of Anhui Medical University in China.
Mechanism Guided Chemoenzymatic Synthesis of Quinolone Alkaloids
New research from Carnegie Mellon University chemists sheds light on how complex natural products could be more effectively made through mechanism guided chemoenzymatic synthesis. The work was published in ACS Catalysis, and was featured as a supplementary journal cover. The cover was designed by former Mellon College of Science Publications Manager and Graphic Designer Rachel Keeney.
Quinoline and quinolone alkaloids are organic compounds from which many potentially valuable products with anti-cancer, antiviral and antibacterial properties can be derived, said Associate Professor of Chemistry Yisong (Alex) Guo.
“People will naturally want to know how you will make more of this,” Guo said.
Naturally, these products are most often found to be synthesized by plants and some microorganisms using a variety of protein catalysts called enzymes. While processes have been previously developed to synthetically create some of the products, the complexity and variety of the functional groups being attached to the molecules in question makes the processes labor intensive and inefficient.
Guo has long worked to better understand and leverage the power of enzymes in synthetic chemical reactions — in research published in the Journal of the American Chemical Society, he and his collaborators looked specifically at the mechanisms of the non-heme-iron oxygenase enzyme AsqJ, which is a critical enzyme in the biosynthesis of quinolone alkaloids.
An important reaction catalyzed by AsqJ is epoxidation, which comprises putting a three-atom ring containing oxygen called an epoxide onto a molecule. This epoxide acts as a functional group for the molecule, allowing it to participate in a variety of chemical reactions including the generation of the core molecular structure of quinolone alkaloids.
In this latest work, Guo and his collaborators built on those mechanistic insights into AsqJ-catalyzed epoxidation by showing how the enzyme could be used to help prepare quinoline and quinolone alkaloid analogues more robustly and efficiently.
“This is the first demonstration of applying this chemoenzymatic synthesis for this family of alkaloids by using non-heme-iron oxygenases,” Guo noted.
With the assistance of molecular dynamics simulations performed by Carnegie Mellon Associate Professor of Chemistry Maria Kurnikova and Visiting Researcher Igor Kurnikov and quantum chemical calculations by Guo, Guo and his longtime collaborator, Wei-chen Chang at North Carolina State University, were able to test a range of potential substrate analogues that AsqJ could catalyze into variants of the quinolone viridicatin. Using these analogue substrates, the researchers then synthesized a wide variety of viridicatin products, including some rarely found in nature.
“Overall, using both enzymes and chemical synthesis with a simple Lewis acid treatment, we can create a library of these man-made compounds at a gram scale,” Guo said.
In addition to Guo, Chan, Kurnikova and Kurnikov, the other researchers on this study were Yijie Tang of Carnegie Mellon; Haoyu Tang of North Carolina State University and Hsuan Jen-Liao and Nei-Li Chan of National Taiwan University.
Computational Chemistry Reveals New Information About Glutamates
The brain’s most prevalent neurotransmitter, glutamate, works differently than previously believed, according to a study from Carnegie Mellon University and Columbia University published in Nature. The study is one of the first to bring together theoretical computational chemistry with experimental biology to establish the structure and function of a molecular machine.
Glutamate is ubiquitous in the human brain, accounting for more than 90% of all synaptic connections. It plays an important role in learning and memory and in disease, including stroke, Alzheimer’s disease and epilepsy.
As a neurotransmitter, glutamate works by binding to receptors on neurons and opening a pore. When ions pass through that pore, they transmit an electrical signal. The brain cells communicate using these electrical signals.
Each receptor on a neuron has four binding sites for glutamate. It was long thought that for each additional glutamate molecule that binds to a neuron, the pore opening would get bigger, resulting in an increase in conductivity. A receptor with one attached glutamate molecule would have a narrow pore and less conductivity and a receptor with four attached glutamate molecules would have the largest pore. The Carnegie Mellon and Columbia team paired cryo-electron microscopy, computational chemistry and machine learning to prove that this wasn’t true.
The Columbia team, led by Alexander Sobolevsky, carried out experiments that allowed them to take cryo-electron microscopy images of glutamate binding to its receptors. They captured images of seven out of 10 possible configurations for glutamate binding to a neuron’s four receptors.
These images alone only allowed the scientists to capture information about the receptors in one isolated space and time. To gain more insight, they turned to computational techniques.
The Carnegie Mellon researchers, led by Associate Professor of Chemistry Maria Kurnikova, took the cryo-electron microscopy images and used them to seed a molecular dynamics simulation that showed how brain cells changed as glutamate bound to each receptor site in each of the different possible configurations. The researchers used high performance computing resources at the Pittsburgh Supercomputing Center, a joint program of Carnegie Mellon and the University of Pittsburgh.
“The molecular dynamics simulations let us look at how a neuron changes over time when glutamate binds to its receptors. The simulation provides data at the level of femtoseconds, giving us a high-resolution picture of activity within the cell,” said Kurnikova.
The researchers then used machine learning to help to make sense of the massive amounts of data created by the simulations. The simulations allowed them to visualize what happens to the pore during different binding patterns — something that couldn’t be seen in the cryo-electron microscopy images alone.
They found that glutamate binds to the four receptor locations in specific patterns, rather than in the previously thought step-wise fashion. They also found that pore size and conductivity didn’t directly correlate with the number of glutamates bound to a neuron. Conductivity was more reliant on the pattern in which they attached to the brain cell.
“This partnership is one of the first to show that computational chemistry simulations can be used to establish structure/function connections in living cells. This opens a huge area of opportunity for research, and computing and machine learning will speed up the experimental process,” said Kurnikova.
This better understanding of the structure and function of glutamate receptors and their activation state could lead to the development of more effective drugs for a variety of diseases that involve glutamate neurotransmitters.
Additional study authors include: Dhilon Patel and Christopher Kottke from Carnegie Mellon and Maria Yelshanskaya from Columbia.