RESEARCH ROUNDUP
HILLER LAB
Carnegie Mellon University researchers have identified a molecule that plays a key role in bacterial communication and infection. Their findings add a new word to pneumococcus’ molecular dictionary and may lead to novel ways to manipulate the bacteria and prevent infection. The findings, from the lab of Associate Professor of Biological Sciences Luisa Hiller, are published in the Oct. 11 issue of PLOS Pathogens.
Organisms worldwide communicate in their own unique ways: humans use words, bees dance, and fireflies glow. Decoding a community’s common language provides the ability to understand and influence the community’s behavior.
What if bacteria also have their own language? If we understood that syntax, could we simply ask the bacteria to stop making us sick?
These questions are at the core of Hiller’s research. Her lab is investigating “bacterial linguistics,” attempting to identify the “words” that bacteria use to communicate. They hope to assemble a dictionary that will give researchers the vocabulary they need to manipulate deadly pathogens.
The Hiller lab is unraveling how bacterial communication contributes to disease and antibiotic resistance, focusing on the bacterium pneumococcus.
Pneumococcus frequently colonizes children and the elderly. It can inhabit the back of the throat without causing any symptoms, but when it spreads to tissues beyond the throat, it can cause mild to severe disease. Pneumococcus is the leading cause of upper respiratory disease and a common cause of pneumonia and pediatric ear infections. Worldwide, it is responsible for more than one million deaths each year.
Pneumococcus forms and thrives in communities called biofilms. The community provides an environment where cells communicate, cooperate and battle. The biofilm also protects the bacteria from antimicrobial interventions and serves as a breeding ground for antibiotic resistance.
In the biofilm, pneumococcus secretes many molecules that appear to be used for communication – telling cells to migrate, multiply and adapt. Despite their potential role in the spread of disease, much about these molecular words is a mystery.
“If we know the meaning of bacterial words, maybe we can remove them from the system or manipulate them so pneumococcus doesn’t become pathogenic,” Hiller said.
In the PLOS Pathogens study, Biological Sciences graduate student Surya Aggarwal identified a previously uncharacterized pneumococcal molecule – a new word – he named BriC. He and his colleagues in the Hiller lab found that BriC plays a critical role in biofilm development by signaling to bacteria to produce more biofilm during competence, a metabolic state that makes multiple cellular functions, including DNA exchange, possible. BriC is researchers’ first clue to understanding how cells in competence interact with other cells to organize themselves into a biofilm.
The study authors also found that in the context of disease, BriC appears to make pneumococcal infections more robust.
“Cells growing within a biofilm are highly recalcitrant to antibiotic treatment” said Aggarwal. “Given this link, BriC assumes greater significance. It could possibly be a target to render bacteria more sensitive to antibiotic treatment.”
In a final twist, the researchers also found that bacteria may also be making semantic changes to their words. They determined that some critically important clinical strains evolved a modification to produce BriC outside of competence. They also produced more BriC, and produced it more often, which enhanced biofilms. The researchers are currently investigating what this variation could mean to the development of disease in children.
“We’re at the very beginning of the dictionary,” said Hiller. “We will continue to dig into the meaning of BriC, and related bacterial words, as a means to manipulate bacterial language and control disease.”
Additional study authors include: Rory Eutsey, Jacob West-Roberts, Wenjie Xu and Aaron Mitchell from Carnegie Mellon University; Arnau Domenech and Jan-Willem Veening from the University of Lausanne; Iman Tajer Abdullah and Hasan Yesilkaya from the University of Leicester and University of Kirkuk; and Hasan Yesilkaya from the University of Leicester. Hiller is also a member of the Center for Excellence in Biofilm Research at the Allegheny Health Network.
The research was funded by the National Institutes of Health (DC011322), the Stupakoff Scientific Achievement Award and Department of Biological Sciences at Carnegie Mellon University.
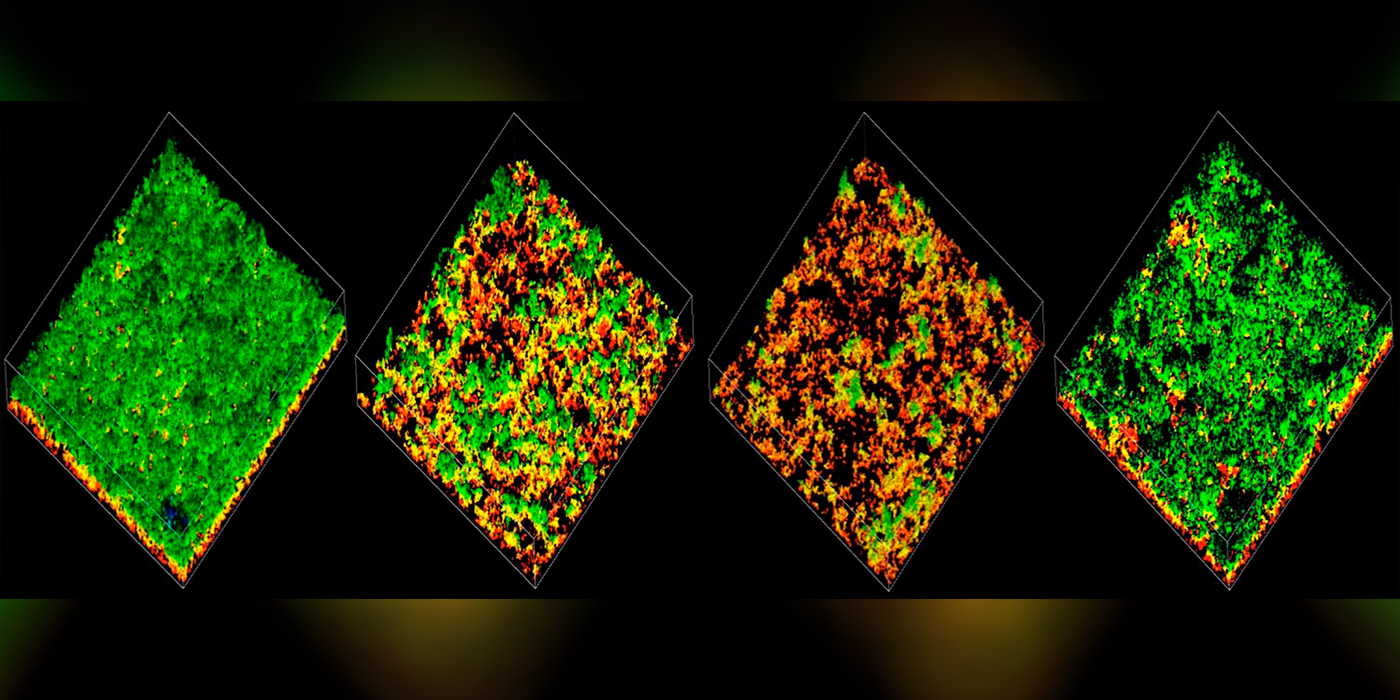
Image: Confocal microscopy images of pneumococcal biofilms colored by depth. Without communication from the BriC peptide, biofilms become less robust.
Related Video
“Intercepting Bacterial Communication to Limit Antibiotic Resistance”
Surya Aggarwal, Three Minute Thesis
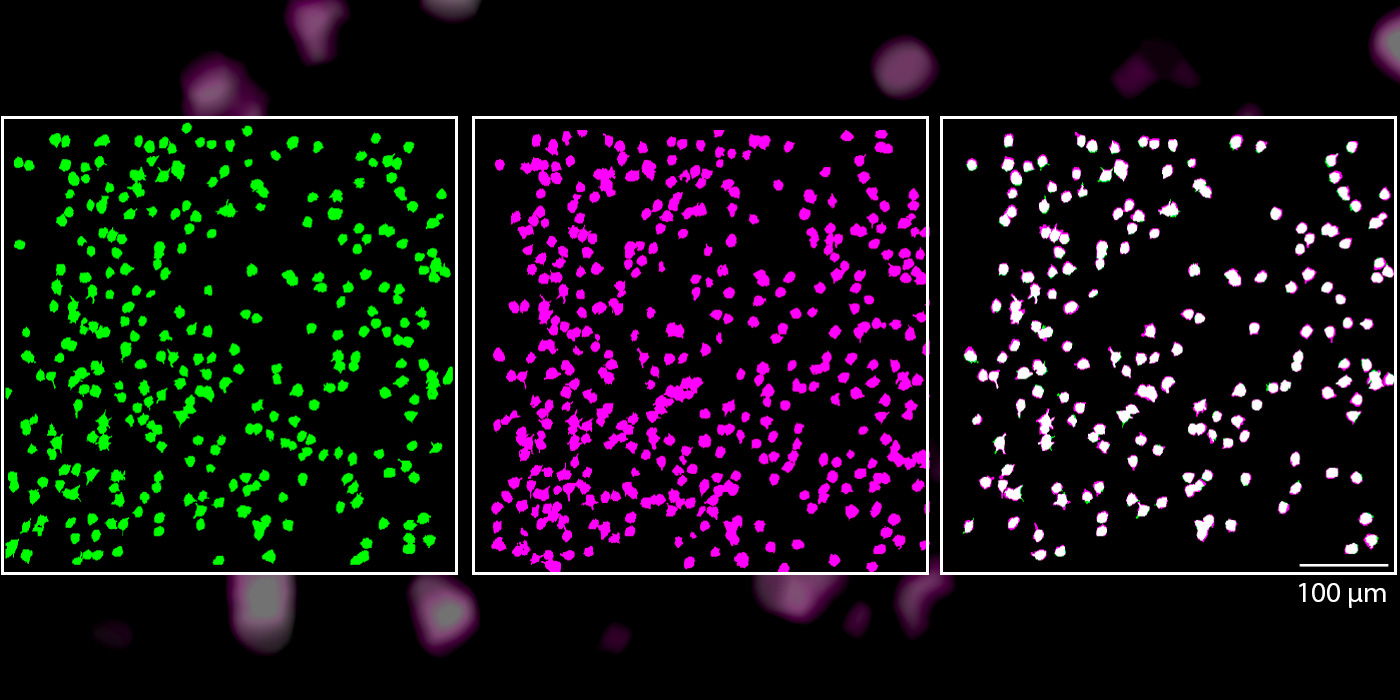
Image: 80% of the tracked neurons were reliably activated by the same oriented lines, indicated that they maintained the same functional role within the brain circuit for days.
KUHLMAN LAB
Single neurons in the brain’s primary visual cortex can reliably detect straight lines, even though the cellular makeup of the neurons is constantly changing, according to a new study by Carnegie Mellon University neuroscientists, led by Associate Professor of Biological Sciences Sandra Kuhlman. The study’s findings, published in Scientific Reports on Oct. 16, lay the groundwork for future studies into how the sensory system reacts and adapts to changes.
Most of us assume that when we see something regularly, like our house or the building where we work, our brain is responding in a reliable way with the same neurons firing. It would make sense to assume that the same would hold true when we see simple horizontal or vertical lines.
“The building our lab is in has these great stately columns,” said Kuhlman. “The logical assumption is that as we approach the building each day our brains are recognizing the columns, which are essentially straight lines, in the same way. Scientifically, we had no idea if this was true.”
While Kuhlman and other scientists believed that this idea of neuronal reliability is a likely hypothesis, they also had reason to believe it might not be the case. The protein components that constitute the cellular makeup of individual neurons continually change over the course of hours or days, which might alter when they respond to a given stimulus. Neither hypothesis had been proven experimentally.
In the case of vision, researchers did know that when we first encounter a stimulus, a group of neurons in the brain’s primary visual cortex respond to the stimulus’ orientation, determining if the stimulus is horizontal, vertical or tilted at an angle. The neurons pass this information deeper into the brain’s visual cortex to the next stage of processing. But they didn’t know which neurons were responding and if the same ones responded each time.
A new imaging technology called two-photon microscopy allowed neuroscientists in Kuhlman’s lab to visualize between 400-600 neurons at once in the primary visual cortex of a mouse model that expresses a fluorescent protein when a neuron is activated. In the experiment, the mouse was shown a sequence pattern of differentially oriented lines — some horizontal, some vertical, and others at angles. These stimuli activated excitatory neurons and caused them to emit a fluorescent signal, which could be seen using the microscope technique.
Over a two-week period, the mice were exposed to the same visual stimuli and researchers measured the response profile of each of the hundreds of neurons. They found that, throughout the study, about 80 percent of the tracked neurons were reliably activated by the same oriented lines. They also reliably remained silent to the same oriented lines. This indicated that they maintained the same functional role within the brain circuit for days.
The researchers were able to test an extensive range of stimuli, including measuring how the neurons responded to lines of varying thickness. They found that some neurons were unstable in how they responded to thickness, while maintaining their original selectivity to line orientation. Kuhlman noted that this indicated that individual neurons can continually encode particular visual features while still being able to adapt to others.
“It was interesting to see plasticity in one feature, but not another,” said Kuhlman. “This gives us a key insight into how our brains may maintain a stable perception of the world while incorporating new information. For example, you want to be able to recognize your building even if slight updates are made, such as if the columns of your building are cleaned. It appears that we can update one aspect of a stimulus feature without completely altering the functional response property of a given neuron.”
The researchers will use this dataset as a control for their next set of studies that aim to see how these neurons respond when there are changes in the visual system, such as while learning a new visual task or following recovery from ocular occlusion.
Additional study authors include: Brian B. Jeon and Steven M. Chase from the CMU Department of Biomedical Engineering and joint Pitt/CMU Center for the Neural Basis of Cognition; Jeffery T. Good from the CMU Department of Biological Sciences; and Alex D. Swain from the University of Pittsburgh Integrative Systems Biology Program.
The study was funded by the National Institutes of Health (EY024678).
McMANUS LAB
New research led by Carnegie Mellon University Associate Professor of Biological Sciences Joel McManus reveals how a group of poorly understood nucleotide sequences in our DNA have a major impact on gene expression.
In our cells, DNA acts as a set of instructions for the production of the protein molecules necessary for life. On its own, however, an instruction manual isn’t very helpful. That’s where RNA comes in, especially messenger RNA (mRNA), which copies and conveys the information encoded in DNA to the cell’s ribosomes. In a process known as translation, these ribosomes scan the copied sequences and build corresponding proteins out of 20 different amino acids.
For each of those 20 amino acids, there are multiple combinations of nucleotides (the familiar letters of A, T, C and G) that can prompt ribosomes to make a particular amino acid. “There’s redundancy in the genetic code,” McManus said. “It turns out this affects the speed of translation.”
This is because when the ribosome is building a protein, it has to wait for the right translation RNA (tRNA) molecule to bring the correct amino acid over to link into the chain. Some nucleotide sequences, known as codons, can take longer to translate than others. This has led biologists to identify “fast” and “slow” codons, and the presence of these can impact how much of a certain protein a cell makes — a cell where the ribosomes work more slowly will end up making less of that protein.
The McManus’ lab’s interest is on short sequences of nucleotides located right before where the ribosomes start translating the mRNA called upstream Open Reading Frames (uORFs), and how those uORFs can repress or amplify the effects of fast and slow codons farther down the mRNA chain, thus affecting the amount of protein production.
“Up to 50 percent of mammalian genes have at least one of these uORFs,” McManus said. “Despite being so common, however, scientists don’t fully understand how they work.”
In a study published in Nucleic Acids Research, McManus and his team set out to answer that question by designing 4,096 different uORFs in yeast and comparing the amount of protein each cell variant made over time.
“Until our study, most people didn’t think these coding sequences in uORFs even mattered,” McManus said when it came to gene expression. “The focus had been only on the amount of uORF translation.”
On the contrary, McManus’ work found that the presence of slow or fast codons within the translated uORFs can have repressive or enhancing effects respectively on the translation process downstream. And the position of the uORFs within the untranslated region of the mRNA can have effects on the protein production.
“It’s not understood what makes some of these uORFs suppress expression and some enhance expression,” McManus noted. “But our work clearly shows that slow and fast codons are important.”
That importance could have major impacts on the differences among cells in our body. Since the amounts of tRNA found in different kinds of human cells vary greatly, so could the effects of the uORFs on translation speed and protein production. “The same sequence in a uORF could be an enhancer in the brain and a repressor in the heart,” McManus said.
Similar differences in enhancement versus repression could also be found when cells are stressed, McManus said, and during the development of cancers. In the long term, McManus plans to start looking at uORFs in human cells to figure out how they work under different conditions, and hopefully build a database of “rules” about how they work.
“We’re excited to look into this more in the future,” McManus said.
Study authors include: Gemma E May, Hunter Kready and Lauren Nazzaro of the Carnegie Mellon Department of Biological Sciences; Yehuda Creeger of the Carnegie Mellon Molecular Biosensor and Imaging Center; Yizhu Lin of the University of California, San Francisco; Peter Spealman of the Center for Genomics and Systems Biology; and Mao Mao of Roche Sequencing Solutions.
The study was funded by the National Institutes of Health (R01 GM121895).
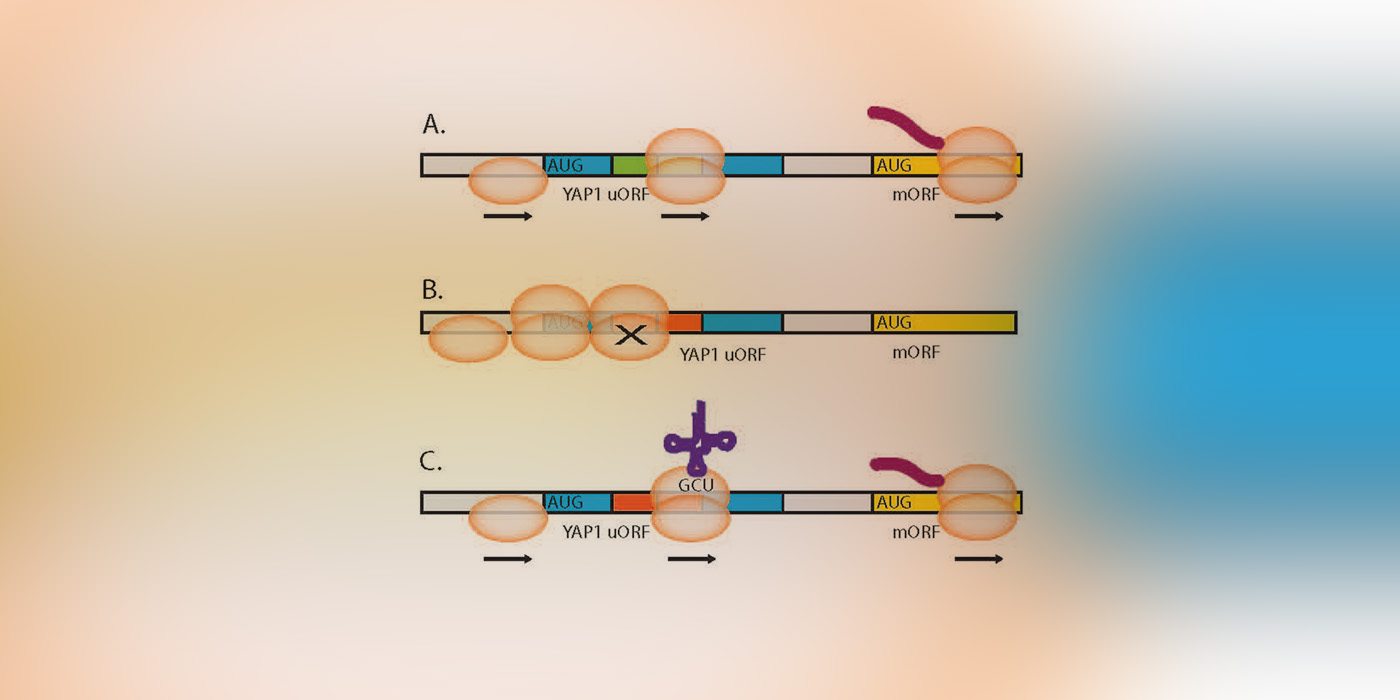
Image: Model of codon impact on uORF regulatory functions.
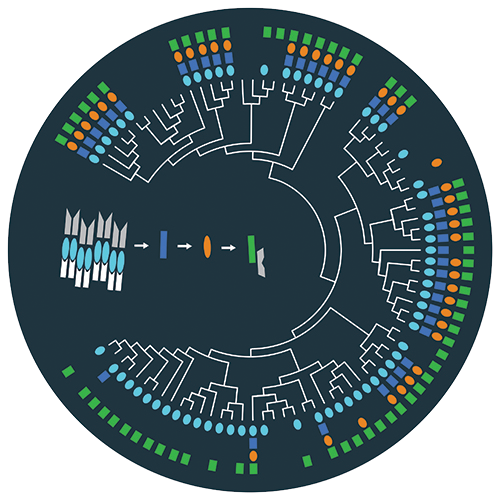
This image illustrates the evolution of the Firmicute sporulation initiation pathway, a compelling example of pathway remodeling.
DURAND LAB
When it comes to changing their passwords, bacteria are just as bad as you and me — maybe even worse. A Carnegie Mellon University research team has found that despite 2.7 billion years of evolution, bacteria are still using the same “password” to initiate the process for making spores. Their findings were published in the September issue of PLOS Genetics.
The Carnegie Mellon researchers, led by the Department of Biological Sciences’ Dannie Durand, used computational and experimental techniques to study how the signaling network that causes Bacilli and Clostridia to form spores has evolved since the two bacteria diverged from a common ancestor 2.7 billion years ago.
Bacteria make spores when times are tough. A protective shell forms around dormant cells to let them withstand harsh conditions like heat, acidity and radiation. Understanding sporulation has implications for many fields, including health care. For example, the spores of C. difficile can survive hand sanitizer, making that bacterium the leading cause of hospital-acquired infections.
Bacteria use “plug-and-play” signaling networks to sense and respond to environmental challenges. A sensor protein recognizes an environmental signal and passes a message to an activator protein, which turns on the appropriate response. Each sensor-activator pair has a specific set of amino acids that act like a password, which ensures that the sensor passes the message to the correct activator.
In the case of the sporulation network, a sensor protein recognizes environmental threats and alerts an activator protein that controls sporulation. The plug-and-play nature of signaling networks makes it easy for bacteria to adapt in a constantly changing world by integrating new sensors into the sporulation network. A sensor that recognizes a new challenge can turn on sporulation, as long as it has the password for the sporulation activator.
Prior studies have shown that in certain well-studied species of Bacillus and Clostridium, the proteins that activate sporulation are very similar, but the signaling networks that deliver the message are not. In Clostridium acetobutylicum, the sporulation signaling network consists of two proteins: the sensor and the activator. In Bacillus subtilis, the network consists of four proteins: the message passes from the sensor through two intermediate proteins before reaching the activator. This raised the question — how did evolution produce different networks that achieve the same results?
It was thought that the common ancestor had the simple two-protein network. Researchers hypothesized that the Bacillar sporulation network gained two proteins over the course of evolution, resulting in the four-protein version we see in Bacilli today. The Clostridia continued to use the two-protein network. This hypothesis was hard to test using standard sequence comparison methods, which are unable to distinguish one signaling network from another.
Durand and colleagues overcame this obstacle by developing a targeted computational approach that combined sequence similarity, protein domain content and neighboring genes to find the genes involved in the sporulation signaling network. When they applied this approach to 28 Bacillar genomes and 56 Clostridial genomes, they found genes encoding four sporulation network proteins in the Bacilli, as expected. Surprisingly, they also found all four genes in many Clostridial genomes.
This overturned the prevailing theory, revealing that the common ancestor must have had the four-protein network. This means that over time some of the Clostridia evolved to use the simpler two-protein network.
“It was surprising, because traditionally we think about evolution going from simple to complex,” said Durand. “But there are more and more examples of evolution going in the other direction, from complex to simple.”
To better understand these changes, study co-author Philip Davidson built artificial four-protein networks in test tubes, each with different combinations of the proteins found in the four-protein sporulation networks. He was able to replace any protein in a Bacillar four-protein sporulation network with the corresponding protein from a Clostridial four-protein network, and vice versa, and the activator still got the message. This shows that Clostridia and Bacilli are still using the same passwords as their ancestor that lived 2.7 billion years ago.
“It’s like your home wireless network. When you first got it, you set up a password and put it in all of your wireless devices. Over the years, you got new computers and smart phones, or had visitors who needed to use the wireless network. If you changed the password, the old devices wouldn’t work, which would be a hassle. So, you continue to use the old password to ensure that everyone could still access the system,” Durand said. “Philip’s experiments show that the Clostridia and the Bacilli get stuck in the same rut when it comes to changing passwords as we do.”
Additional study authors include: Rory Eutsey, Brendan Redler and N. Luisa Hiller, Carnegie Mellon Department of Biological Sciences; and Michael T. Laub, Department of Biology, Massachusetts Institute of Technology.
The study was funded by the Human Frontiers Science Program and National Science Foundation (DBI-1262593). Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Laub is an investigator of the Howard Hughes Medical Institute.